Scientists store 900,000GB of data in 1 gram of E. coli bacteria

Doesn’t seem possible? Well, scientists have been working on using proteins, bacteria and other organic material as storage media for a while now, and if it looks like all those efforts are bearing fruit now, it doesn’t make it any more unlikely! Calling it ‘bioencryption by recombination’, a team of scientists from Chinese University of Hong Kong (CUHK) have figured out how to store and en/decrypt data onto living bacteria cells.
 Survey
SurveyThese efforts are part of the CUHK’s submission to iGEM (International Genetically Engineered Machine) 2010 contest, and its mission statement reads:
CUHK iGEM 2010 team is formed by a group of undergraduates and instructors from the Chinese University of Hong Kong. Our project is to create a brand new biological cryptography system. We harness the incredible adaptability of simple organisms in the tortured environment to make sure that the message stored can be left undisturbed regardless of any environmental changes.
[RELATED_ARTICLE]As you can infer, the aim of the project is not just to create an information dense storage medium, but also to make it extremely resistant to hacking and environmental damage, which most current solutions are especially affected by. You can download their presentation (PDF) from here. In essence, the team sought to make bacteria data storage and encryption feasible in the real world, which previously returned very low and impractical data density figures. Now, they’ve managed to squeeze more than 931,322GB of data onto 1 gram of bacteria (specifically a DH5-alpha strain of E.coli, chosen for its extracted plasmid DNA size) by creating a massively parallel bacterial data storage system. Compared to 1 to 4GB per gram data density of conventional media, the 900,000GB per gram figure the team has returned is truly astounding.
Taking the dream one step closer to industrial reality, the team has developed data proof-read/correction and random access modules, in addition to an encryption module, all using site-specific recombination of the inversion type, specifically, R64 Shufflon-Specific Recombinase, a type of Rci-mediated recombination. In essence, the team has transferred information onto DNA, and the encoding method to do this has been explained below:
A translation table would first need to be constructed by the client, the extended ASCII table with 256 characters were used as standard in here. It is not difficult to identify DNA as a naturally referred as a quaternary numeral system, With the DNA base adenosine representing the number “0”, thymine representing “1”, cytosine representing “2” and guanine representing “3”, we are essentially encoding the 256 characters with this base-4 numeral system.
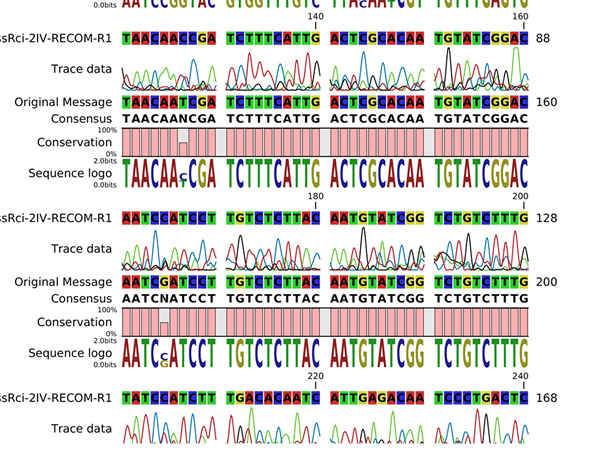
Before the DNA is synthesised, the resultant code/DNA information is compressed using a combination of Huffman coding and LZ77 algorithm, allowing for reduced “homopolymer and repetitive regions”, and, more information to coded into less units.