How to use your PC to contribute towards Coronavirus research
There are two distributed computing projects that are working on treatment research for the novel Coronavirus, Folding@Home and Rosetta@Home.
One of the ways in which you can help with the ongoing pandemic is to use excess PC resources towards distributing computing projects. These projects investigate the structure of proteins, to find possible therapies and treatments. There are two distributed computing projects that are working on treatment research for the novel Coronavirus, Folding@Home and Rosetta@Home.
Method 1: Contribute to Folding@Home
Folding@home is a project that studies the structure of proteins, how the various molecules move, and how they fold up to realise their final forms. This research allows scientists to come up with treatment therapies, based on the geometries they discover of the proteins. Apart from the Coronavirus, the project has been used to investigate treatments for Cancer and Alzheimer’s disease as well. The software has been developed by researchers at Stanford University.
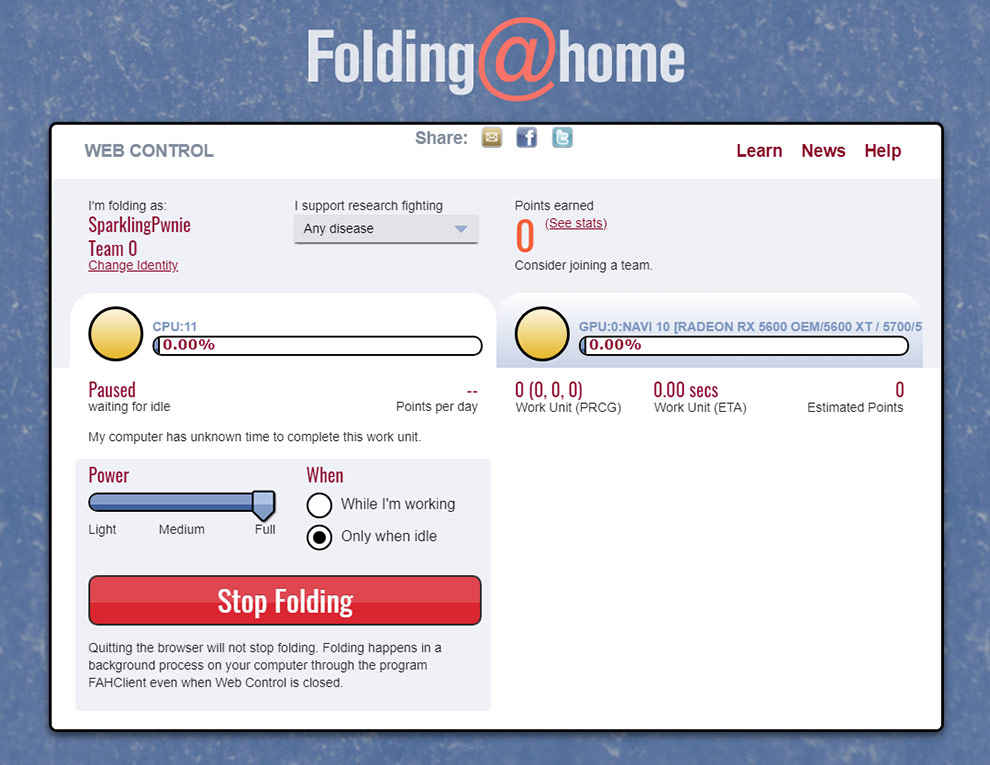
The Folding@Home interface
Step 1: Install Folding@Home
If you have Windows, then you can install Folding@Home from this link. In case you have a Mac or using the Linux OS, you can find the relevant package on the alternative downloads page. Once the installer has been downloaded, run it. The express installer just goes ahead and installs the software in the C drive, but the advanced installation option lets you change the location. The software will then ask permission to be allowed through the firewall, allow for both Private and Public networks if the software is being installed on office computers. An Android client was developed in collaboration with Sony, it has been open sourced, and there are APKs available, but the client is not on the Play Store.
Step 2: Setup the software
The software will use your default browser as an interface, but it is actually a local client. You will need to come back to this page to interact with the software, although it is running in the background on your PC. A handy way to do this is to bookmark the page, which is something Folding@Home recommends as well. You can now set up an identity, which is a profile that is used to track your contributions. You can also join teams and earn points, depending on how much computing resources you have donated. However, users also have the choice of skipping this step, and contributing to the project anonymously. Now, you can choose to secure your profile with a passkey. A passkey is a 32 char hexadecimal string, something like “1879901cf9b591e74569a646561edd41” which is too long to remember obviously. You can generate one here. Select "32" for "How many digits?" and select “1” in the “How many results?” field, then feed that string into the passkey field on the Folding@Home local client. Remember to email the correct passkey to yourself, as Folding@Home does not collect any information such as email id or phone number to reclaim a profile that you have lost the passkey to.
Step 3: Start Folding
Now, you can start setting up the software to decide the amount of resources dedicated to the folding. The power slider can be adjusted from Light to Full. You can also choose to fold proteins only when the computer is idling. Finally, pick the disease you want to research. At the moment, all the research related to Coronavirus are bunched under the “Any Disease” category. That’s it, you are now contributing to Coronavirus research. Do note that your computer will see a performance hit, and that you have to stop folding before closing the web interface. If you are leaving your machine on and idling, then remember to start folding through the local web client. More information about the project and how you can contribute is available on the Folding@Home website.
Method 2: Contribute to Rosetta@Home
Similar to Folding@Home, Rosetta@Home is another project that works towards investigating the structures of proteins, and analysing them for any possible treatments and therapies. A number of important Coronavirus proteins have already been modelled through the project, and a list of these models uncovered through Rosetta@Home volunteers is available here. The Rosetta@Home uses the Berkeley Open Infrastructure for Network Computing (BOINC) software.
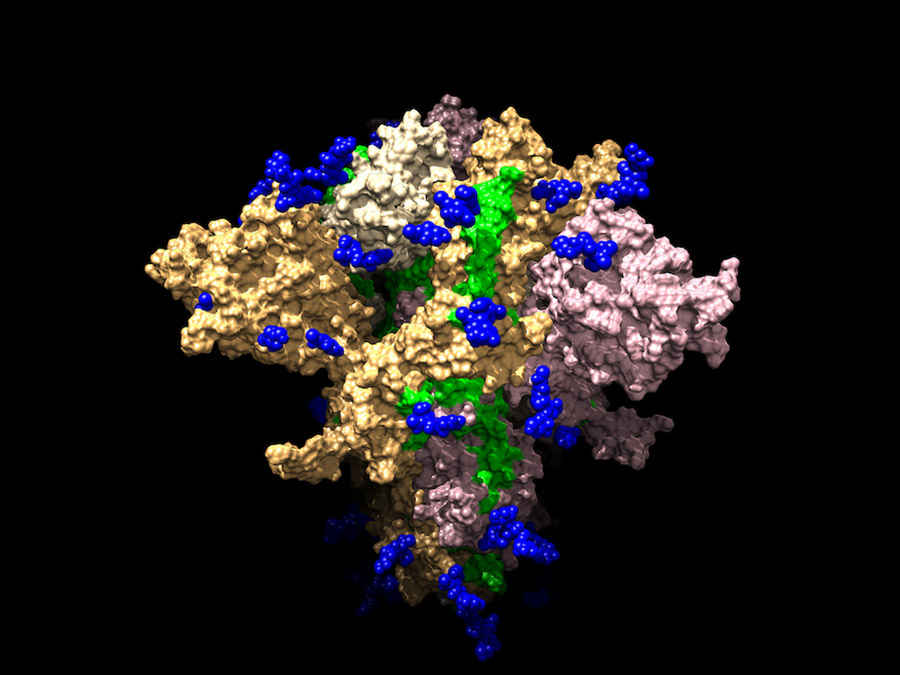
Model of the SARS-CoV-2, also called 2019-nCov Spike protein, that causes COVID-19. This structure was determined through the help of Rosetta@Home.
Credit: ssgcid
Step 1: Download and install BOINC
There are two ways to go about this. There are two options, BOINC+VirtualBox, and just BOINC, which can be downloaded here. The first option, which is recommended, allows you to set up a virtual PC with a set amount of resources dedicated to the science projects. It also isolates the software from your system. The other install allows you to install BOINC natively on your PC. You can use Advanced Installation to customize the location. There is also an Android client for BOINC, which is available on the Play Store. After installation, run the BOINC manager.
Step 2: Add the Rosetta@Home project
In the BOINC manager, you will see a number of categories, and particular projects. Each of these projects have a description, so that you are aware of what you are contributing computer resources to. The Rosetta@Home project is available under the Biology and Medicine category. Select the project, and click on “Next”. At this point, the software will communicate with the project servers, and ask you to create a new account. Rosetta@Home asks for an email id and a password. Enter these in, and select Next. If you have a problem signing up ,you can do so through the Rosetta@Home website, then login to the client on BOINC. The Rosetta@Home project is now added to the BOINC client.
Step 3: Start contributing
Now, make sure that in the Firewall rules, the BOINC client, the BOINC manager, and the BOINC command softwares are allowed to communicate with their servers. In the Computing Preferences section, you can tweak a number of options. If you have installed the software on a laptop, BOINC can be configured to stop working when the device is not plugged in. You can also specify how much of CPU time to dedicate to the computing. You can also specify how much space downloaded project files should occupy, when they should be downloaded, and schedule the time when the projects should run. The software can also be configured to detect a lack of keyboard/mouse activity, after which it starts running the projects. Note that you can contribute to multiple science projects at the same time in BOINC, by adding them one by one.
Cover Image credit: Alissa Eckert, MS; Dan Higgins, MAMS, CDC
Aditya Madanapalle
Aditya Madanapalle, has studied journalism, multimedia technologies and ancient runes, used to make the covermount DVDs when they were still a thing, but now focuses on the science stories and features. View Full Profile